BH11.11/統合検索とRDFからの表データ生成/uniprot sparql
提供:TogoWiki
Names and origin
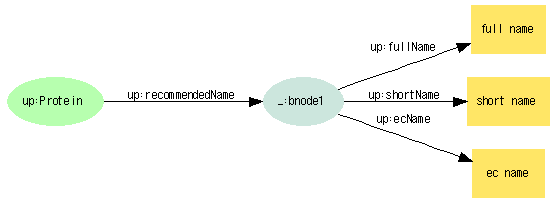
Protein names
Recommended name
Full name
SELECT ?id ?recommended_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?recommended_name_node <http://purl.uniprot.org/core/fullName> ?recommended_name . ?id <http://purl.uniprot.org/core/recommendedName> ?recommended_name_node }
Short name(s)
SELECT ?id ?short_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?recommended_name_node <http://purl.uniprot.org/core/shortName> ?short_name . ?id <http://purl.uniprot.org/core/recommendedName> ?recommended_name_node }
EC
SELECT ?id ?ec FROM <http://bh.dbcls.jp/DAV> WHERE { ?recommended_name_node <http://purl.uniprot.org/core/ecName> ?ec . ?id <http://purl.uniprot.org/core/recommendedName> ?recommended_name_node }
Alternative name(s)
Full name
SELECT ?id ?alternative_names FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_names_node <http://purl.uniprot.org/core/fullName> ?alternative_names . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_names_node }
Short name(s)
SELECT ?id ?short_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/shortName> ?short_name . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
EC
SELECT ?id ?ec FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/ecName> ?ec . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
Allergen
SELECT ?id ?allergen FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/allergenName> ?allergen . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
Biotech
SELECT ?id ?biotech FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/biotechName> ?biotech . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
CD_antigen
SELECT ?id ?cd_antigen FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/cdAntigenName> ?cd_antigen . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
INN
SELECT ?id ?inn FROM <http://bh.dbcls.jp/DAV> WHERE { ?alternative_name_node <http://purl.uniprot.org/core/internationalNonproprietaryName> ?inn . ?id <http://purl.uniprot.org/core/alternativeName> ?alternative_name_node }
Gene names
Name
SELECT ?id ?gene_names FROM <http://bh.dbcls.jp/DAV> WHERE { ?gene_names_node <http://www.w3.org/2004/02/skos/core#prefLabel> ?gene_names . ?id <http://purl.uniprot.org/core/encodedBy> ?gene_names_node }
Synonyms
SELECT ?id ?alt_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?gene_names_node <http://www.w3.org/2004/02/skos/core#altLabel> ?alt_name . ?id <http://purl.uniprot.org/core/encodedBy> ?gene_names_node }
Ordered Locus Names
SELECT ?id ?locus_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?encoded_by <http://purl.uniprot.org/core/locusName> ?locus_name . ?id <http://purl.uniprot.org/core/encodedBy> ?encoded_by }
ORF Names
SELECT ?id ?orf_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?encoded_by <http://purl.uniprot.org/core/orfName> ?orf_name . ?id <http://purl.uniprot.org/core/encodedBy> ?encoded_by }
Encoded on
SELECT ?id ?organelle ?label
WHERE {
?encoded_in <http://www.w3.org/2000/01/rdf-schema#label> ?label .
?encoded_in <http://www.w3.org/2000/01/rdf-schema#subClassOf> ?organelle .
?id <http://purl.uniprot.org/core/encodedIn> ?encoded_in
}
Organism
SELECT ?id ?organism FROM <http://bh.dbcls.jp/DAV> WHERE { ?taxonomy_id <http://purl.uniprot.org/core/scientificName> ?organism . ?id <http://purl.uniprot.org/core/organism> ?taxonomy_id }
include=yes でRDFを取得した場合。オプションを付けずにRDFを取得した場合にはtaxonomy_idのみ取れる(その他の情報はtaxonomyのグラフから取る)
Taxonomic identifier
SELECT ?id ?taxonomic_identifier FROM <http://bh.dbcls.jp/DAV> WHERE { ?id <http://purl.uniprot.org/core/organism> ?taxonomic_identifier }
Taxonomic lineage
Virus host
SELECT ?id ?host FROM <http://bh.dbcls.jp/DAV> WHERE { ?taxonomy_id <http://purl.uniprot.org/core/host> ?host . ?id <http://purl.uniprot.org/core/organism> ?taxonomic_identifier }
Protein attributes
Sequence length
SELECT ?id ?sequence FROM <http://bh.dbcls.jp/DAV> WHERE { ?seq <http://www.w3.org/1999/02/22-rdf-syntax-ns#value> ?sequence . ?id <http://purl.uniprot.org/core/sequence> ?seq FILTER regex (?id, "uniprot") }
で返ってくる文字列の長さ
Sequence status
SELECT ?id ?value FROM <http://bh.dbcls.jp/DAV> WHERE { ?seq <http://purl.uniprot.org/core/fragment> ?value . ?id <http://purl.uniprot.org/core/sequence> ?seq }
- if ?value == "single" then "Fragment" elsif ?value == "multiple" then "Fragments" elsif ?value == nil then "Complete"
Sequence processing
Protein existence
SELECT * FROM <http://bh.dbcls.jp/DAV> WHERE { ?id <http://purl.uniprot.org/core/existence> ?existence }
- 5 types of evidence for the existence: 'Evidence at protein level', 'Evidence at transcript level', 'Inferred from homology', 'Predicted', 'Uncertain'
General annotation (Comments)
SELECT ?id ?type ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> ?type . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
- ?type からどの項目のアノテーションか判別可能
Function
SELECT ?id ?type ?function_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?function_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Function_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Catalytic activity
SELECT ?id ?catalytic_activity_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?catalytic_activity_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Catalytic_Activity_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Cofactor
SELECT ?id ?cofactor_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?cofactor_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Cofactor_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Enzyme regulation
SELECT ?id ?enzyme_regulation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?enzyme_regulation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Enzyme_Regulation_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Biophysicochemical properties
Subunit structure
SELECT ?id ?subunit_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?subunit_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Subunit_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Pathway
SELECT ?id ?pathway_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?pathway_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Pathway_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Subcellular location
SELECT ?id ?location ?subcellular_location_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?subcellular_location_annotation_comment . ?located_in ?p ?location . ?annotation <http://purl.uniprot.org/core/locatedIn> ?located_in . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Subcellular_Location_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Tissue specificity
SELECT ?id ?tissue_specificity_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?tissue_specificity_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Tissue_Specificity_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Developmental stage
SELECT ?id ?developmental_stage_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?developmental_stage_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Developmental_Stage_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Induction
SELECT ?id ?induction_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?induction_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Induction_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Domain
SELECT ?id ?domain_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?domain_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Domain_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Post-translational modification
SELECT ?id ?ptm_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?ptm_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/PTM_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
RNA editing
Mass spectrometry
Polymorphism
Involvement in disease
Disruption phenotype
Allergenic properties
Toxic dose
Biotechnological use
Pharmaceutical use
Miscellaneous
Sequence similarities
SELECT ?id ?similarity_annotation_comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?similarity_annotation_comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Similarity_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Caution
Sequence caution
Ontologies
Keywords
Biological process
Cellular component
Coding sequence diversity
Developmental stage
Disease
Domain
Ligand
Molecular function
Post-translational modification
Technical term
Gene Ontology (GO)
Biological process
Cellular component
Molecular function
Binary interactions
SELECT ?id ?interact_with ?interact_entry ?num_experiments ?participant ?xeno FROM <http://bh.dbcls.jp/DAV> WHERE { ?participant1 <http://www.w3.org/2000/01/rdf-schema#label> ?interact_with . ?participant1 <http://www.w3.org/2002/07/owl#sameAs> ?interact_entry . ?interaction <http://purl.uniprot.org/core/xeno> ?xeno . ?interaction <http://purl.uniprot.org/core/experiments> ?num_experiments . ?interaction <http://purl.uniprot.org/core/participant> ?participant1 . ?interaction <http://purl.uniprot.org/core/participant> ?participant . ?id <http://purl.uniprot.org/core/interaction> ?interaction }
- with, entry, #Exp., IntAct, Notes の5項目で一セットの情報
Alternative products
General information
Isoform-specific information
Identifier
Name and synonym(s)
Description of the sequence differences
Additional comments
Sequence annotation (Features)
Molecule processing
Initiator methionine
Signal
Transit peptide
- Mitochondrion, Apicoplast, Chromoplast, Chloroplast, Cyanelle, Thylakoid, Amyloplast, Peroxisome, Glyoxysome, Hydrogenosome
Propeptide
SELECT ?id ?begin ?end ?comment ?annotation FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Propeptide_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Chain
SELECT ?id ?begin ?end ?comment ?annotation FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Chain_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Peptide
Regions
Topological domain
Transmembrane
Intramembrane
Domain
Repeat
Calcium binding
Zinc finger
DNA binding
Nucleotide binding
SELECT ?id ?begin ?end ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/NP_Binding_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Region
SELECT ?id ?begin ?end (?comment AS ?merge)
WHERE { OPTIONAL {
?range <http://purl.uniprot.org/core/end> ?end .
?range <http://purl.uniprot.org/core/begin> ?begin .
?annotation <http://purl.uniprot.org/core/range> ?range .
?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment .
?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Region_Annotation> .
?id <http://purl.uniprot.org/core/annotation> ?annotation .
}}
Coiled coil
Motif
SELECT ?id ?begin ?end ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Motif_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Compositional bias
Sites
Active site
Metal binding
Binding site
Site
Amino acid modifications
Non-standard residue
Modified residue
SELECT ?id ?begin ?end ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Modified_Residue_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Lipidation
SELECT ?id ?begin ?end ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Lipidation_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Glycosylation
Disulfide bond
Cross-link
Natural variations
Alternative sequence
SELECT ?id ?begin ?end ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Alternative_Sequence_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Natural variant
Experimental info
Mutagenesis
SELECT ?id ?begin ?end ?substitution ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://purl.uniprot.org/core/substitution> ?substitution . ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Mutagenesis_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Sequence uncertainty
Sequence conflict
Non-adjacent residues
Non-terminal residue
Secondary structure
Helix
SELECT ?id ?begin ?end FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Helix_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Turn
SELECT ?id ?begin ?end FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Turn_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Beta strand
SELECT ?id ?begin ?end FROM <http://bh.dbcls.jp/DAV> WHERE { ?range <http://purl.uniprot.org/core/end> ?end . ?range <http://purl.uniprot.org/core/begin> ?begin . ?annotation <http://purl.uniprot.org/core/range> ?range . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://purl.uniprot.org/core/Beta_Strand_Annotation> . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
Sequences
Sequence
SELECT ?id ?sequence FROM <http://bh.dbcls.jp/DAV> WHERE { ?seq <http://www.w3.org/1999/02/22-rdf-syntax-ns#value> ?sequence . ?id <http://purl.uniprot.org/core/sequence> ?seq FILTER regex (?id, "uniprot") }
Length
- ↑Sequenceの結果返ってくる文字列の長さ
Mass (Da)
SELECT ?id ?mass FROM <http://bh.dbcls.jp/DAV> WHERE { ?seq <http://purl.uniprot.org/core/mass> ?mass . ?id <http://purl.uniprot.org/core/sequence> ?seq FILTER regex (?id, "uniprot") }
References
Number
Title
Author(s) names
Reference information
Cross-references
SELECT ?id ?o ?pmid FROM <http://bh.dbcls.jp/DAV> WHERE { ?o <http://www.w3.org/2004/02/skos/core#exactMatch> ?pmid . ?id <http://purl.uniprot.org/core/citation> ?o FILTER regex (?pmid, "pubmed") }
http://purl.uniprot.org/pubmed/#{pmid}
Citation content (Cited for)
Sequence origin
Web resources
Cross-references
SELECT ?id ?xref_id FROM <http://bh.dbcls.jp/DAV> WHERE { ?id <http://www.w3.org/2000/01/rdf-schema#seeAlso> ?xref_id }
http://purl.uniprot.org/#{DB_name}/#{id}
- total 145 DBs
2D gel databases
-
2DBase-Ecoli,ANU-2DPAGE,COMPLUYEAST-2DPAGE,Cornea-2DPAGE,DOSAC-COBS-2DPAGE,OGP,PHCI-2DPAGE,PMMA-2DPAGE,Rat-heart-2DPAGE,REPRODUCTION-2DPAGE,Siena-2DPAGE,SWISS-2DPAGE,UCD-2DPAGE,World-2DPAGE,Aarhus/Ghent-2DPAGE,ECO2DBASE - 16 DBs (0 DB)
3D structure databases
- DisProt,
HSSP,ModBase, PDB,PDBsum,ProteinModelPortal,SMR - 7 DBs (2 DBs)
Enzyme and pathway databases
- BioCyc, BRENDA,
ENZYME, Pathway_Interaction_DB, Reactome - 5 DBs
Family and domain databases
- Gene3D, HAMAP, InterPro, KO, PANTHER, Pfam, PIRSF, PRINTS, ProDom, PROSITE, SMART, SUPFAM, TIGRFAMs
- 13 DBs (13 DBs)
Gene expression databases
-
ArrayExpress,Bgee, CleanEx,Genevestigator, GermOnline - 5 DBs
Genome annotation databases
-
CMR, Ensembl, EnsemblBacteria, EnsemblFungi, EnsemblMetazoa, EnsemblPlants, EnsemblProtists, GeneID, GenomeReviews, KEGG, NMPDR, TIGR, VectorBase, UCSC, PATRIC - 15 DBs (14 DBs)
Ontologies
- GO
- 1 DB
Organism-specific databases
- AGD, ArachnoServer, CGD, ConoServer, CTD, CYGD, dictyBase, EchoBASE, EcoGene, euHCVdb, EuPathDB, FlyBase,
GenAtlas, GeneCards, GeneDB_Spombe, GeneFarm, GenoList,Gramene, H-InvDB, HGNC, HPA,HUGE, LegioList, Leproma, MaizeGDB, MGI,Micado, MIM, neXtProt, Orphanet, PharmGKB, PseudoCAP, RGD,Rouge, SGD, TAIR, TubercuList, WormBase, Xenbase, ZFIN - 40 DBs
Phylogenomic databases
- eggNOG, GeneTree, HOGENOM, HOVERGEN,
InParanoid, OMA, OrthoDB,PhylomeDB, ProtClustDB - 9 DBs
Polymorphism databases
- dbSNP (in Natural variations), DMDM
- 2 DBs (1DB)
Protocols and materials databases
-
SBKB - 1 DB (0 DB)
Proteomic databases
-
PeptideAtlas,PRIDE,ProMEX - 3 DBs (0 DB)
Protein-protein interaction databases
- DIP,
IntAct, MINT,STRING - 4 DBs (2 DBs)
Protein family/group databases
- Allergome,
AraC-XylS, CAZy,GPCRDB,IMGT, MEROPS, PeroxiBase, PptaseDB, REBASE, TCDB - 10 DBs (7 DBs)
PTM databases
-
GlycoSuiteDB,PhosphoSite,PhosSite - 3 DBs (0 DB)
Sequence databases
- EMBL/GenBank/DDBJ, IPI, PIR, RefSeq, UniGene
- 7 DBs (5 DBs)
Other
-
BindingDB, DrugBank, NextBio,PMAP-CutDB,ProtoNet,SOURCE - 6 DBs (2 DBs)
Entry information
Entry name
SELECT ?id ?entry_name FROM <http://bh.dbcls.jp/DAV> WHERE { ?id <http://purl.uniprot.org/core/mnemonic> ?entry_name FILTER regex(?id, "http://purl.uniprot.org/uniprot") }
Accession
- UniProt ID itself
Entry history
Entry status
SELECT ?id ?reviewed FROM <http://bh.dbcls.jp/DAV> WHERE { ?id <http://purl.uniprot.org/core/reviewed> ?reviewed FILTER regex(?id, "http://purl.uniprot.org/uniprot") }
- 1=reviewed, 0=unreviewed
General annotation and Sequence annotation
SELECT ?id ?type ?comment FROM <http://bh.dbcls.jp/DAV> WHERE { ?annotation <http://www.w3.org/2000/01/rdf-schema#comment> ?comment . ?annotation <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> ?type . ?id <http://purl.uniprot.org/core/annotation> ?annotation }
- typeから、どの項目のアノテーションか判別可能